Since 2006, our well-experienced team has been delivering molecular profiles, such as affinity and kinetic characterisation, as well as mechanistic evaluation insights for a wide range of different proteins, drug target types and purposes.
Examples of services
Our service offering include, but is not limited to:
Small molecule protein interaction analysis
Kinetic and mechanistic characterisation
Controlling the kinetics to increase chances of clinical efficacy
Benefits
The efficacy of inhibiting a target is dependent on the rate of target inactivation and the residence time of the ligand. Therefore, two different ligands with identical affinity can differ significantly in clinical efficacy due to differences in kinetics. Experiments can often be designed to be qualitative or quantitative.
Gaining control over the association and dissociation rate of your ligand can provide a competitive advantage. In addition, the discovery of ligands with unexpected kinetics can provide an inventive step, making your compounds patentable.
By varying the assay conditions, the mechanism of ligand binding can be scrutinized.
Technical Details
The analysis of protein-ligand interactions is performed by SPR biosensor-based experiments. This entails the immobilisation of the target protein onto a sensor chip, followed by injection of a concentration series of reference and test compounds.
Analysis of the response with respect to injection time allows for the assignment of the interaction mechanism and the determination of the corresponding rate constants as well as dissociation constants.
Public examples of work:
Nimbus Therapeutics, Cambridge, MA, USA (Wei et. al. Nature, 2019, PMID30944472)
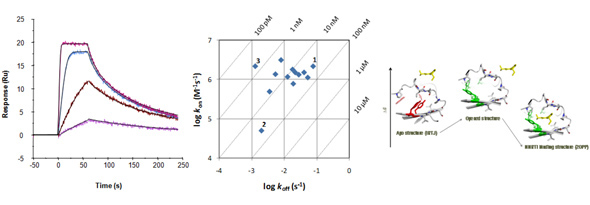
Examples of kinetic traces of a kinase inhibitor binding via a simple direct fit mechanism (left). Kinetic data can be shown in kon/koff plots (middle). Compounds 1 and 2 are equipotent in terms of affinity but show differences in kinetics. By increasing on-rate for 2 or by decreasing off-rate for 1, the potency of 3 could be reached. Scheme to the right shows a more complex mechanistic mechanism (selected fit) illustrated by structures of HIV-1 RT.
Antibody interaction analysis
Understanding antibody kinetics and selectivity
Control the kinetics to increase chances of clinical efficacy
Benefits
SPR biosensor-based technology has several advantages for antibody characterisation over conventional methods (such as enzyme immunoassays and radioimmunassay). No labelling is required, sample consumption is low, the method is fast and the output is information rich. Flexible experimental design can provide information about such diverse parameters as interaction model, affinity, rate constants, temperature- and pH dependency as well as epitope specificity.
Technical Details
In epitope mapping, epitope specificity is determined by testing the ability of pairs of monoclonal antibodies (mAbs) to bind simultaneously to an antigen. Typically, a first mAb is immobilised onto the chip surface and then utilised to capture an antigen of interest. A second mAb is subsequently flowed over the antigen surface. If the second mAb is directed towards an epitope distinct from that of the first mAb (non-competitive), it will bind, whereas if it is directed towards an epitope closely related to the epitope of the first mAb it will interfere with binding (competitive).
Other service offerings include standard affinity and interaction kinetics determination, analysis of Fc γ receptor panel interactions, and concentration assessments.
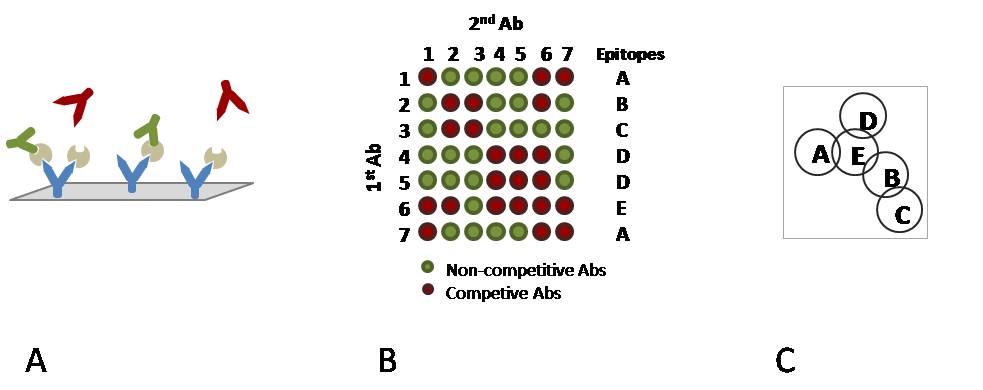
A) Immobilised mAbs on a chip surface with captured antigen and two types of ”secondary” mAbs. Green mAbs are non-competitive binders whereas red mAbs are competitive.
B) A reactivity matrix showing the binding ability of pairs of mAbs to an antigen along with epitope pattern definitions.
C) A diagram representing the relationship between epitopes. Overlapping epitopes are competitive, non-overlapping epitopes are non-competitive.
Protein / protein interaction analysis
Quantify macromolecule interactions
Understand what drives binary and ternary protein complex formation
Benefits
Molecular interactions are fundamental for all life. Proteins build up binary and higher order complexes. At which rate these complexes assemble and fall apart is central for function and thus biologic effect. For ternary (and higher order) complexes, the cooperativity of binding determines the propensity of complex formation.
Technical details
One protein is tethered to the hydrogel of a biosensor chip through an antibody or some other affinity handle, or linked covalently to the gel matrix. A high protein purity is not crucial for non-covalent capture techniques. Concentration series of the liganding protein is injected and the interaction is recorded in real-time, without the use of any labels.
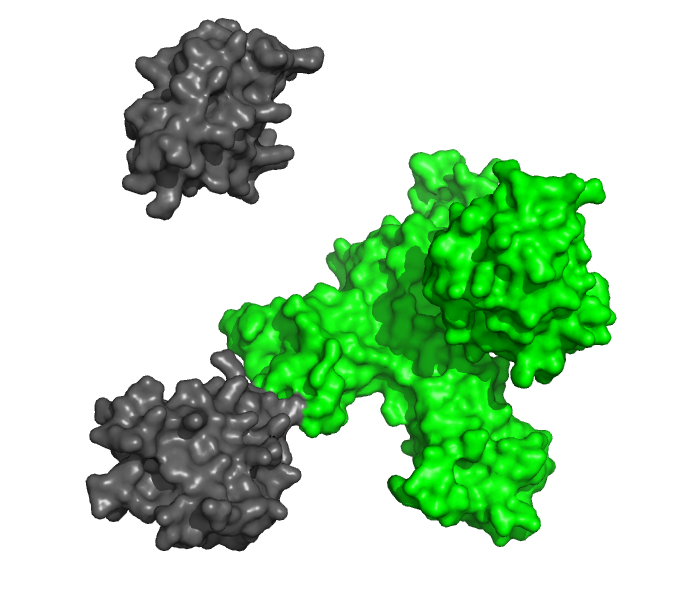
Study the interactions between protein ligands and their receptors in real time, without any potentially interfering labels.
Analysis of extreme kinetics
Quantify long residence times to rank late stage lead-optimization ligands
The Extended Rate Analysis (ExtRA™) method
Benefits
In the late stages of lead optimization, it is common that compounds dissociate very slowly from their target. For such interactions, steady-state approaches are not useful and standard kinetic methods can only reliably quantify dissociation rates faster than ~10-4 s-1 (Önell & Andersson, J. Mol. Recogn. 2005;18:307-317).
Technical details
By a innovative experimental design, the dissociation rate of very high affinity ligands can be determined. This provides a tool to evaluate and further optimize late stage lead-optimization compounds.
The ExtRA™ method has been successfully applied in several partner projects, e.g. with Constellation Pharmaceuticals, Cambridge, MA USA (see Stuckey et. al. PMID33524394).
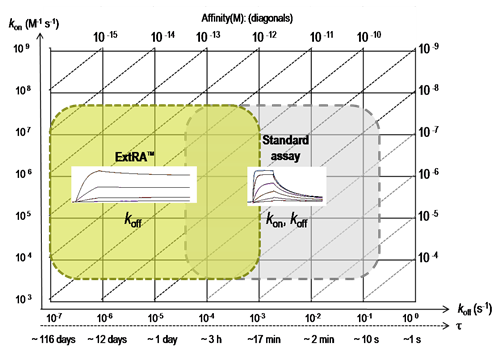
ExtRA™ range for quantification of dissociation rate constants (koff) or residence times (τ), compared to standard SPR-based assays.
Deliverables
A reliable measurement of affinity (i.e. the KD value) for sub nM interactions
Kinetic dissociation rates (koff) down to 10-7 s-1
Determine residence times of >100 days
Mechanistic interpretation
Stay Updated
Sign up for the Beactica newsletter to receive our latest news and updates
